interaction
Macromolecules, such as proteins, perform their functions by dynamically changing their interaction partners in the heterogeneous cellular environment. Taking advantage of the strength of NMR, which can analyze interactions at atomic resolution even in heterogeneous systems, we have elucidated the functional roles of specific as well as seemingly unnecessary non-specific interactions of macromolecules. The interaction analysis by NMR is useful for clarifying the functional role of biomolecular interaction and for structure-guided optimization of bioactive compounds.
Dynamic ligand optimization by the FCT method

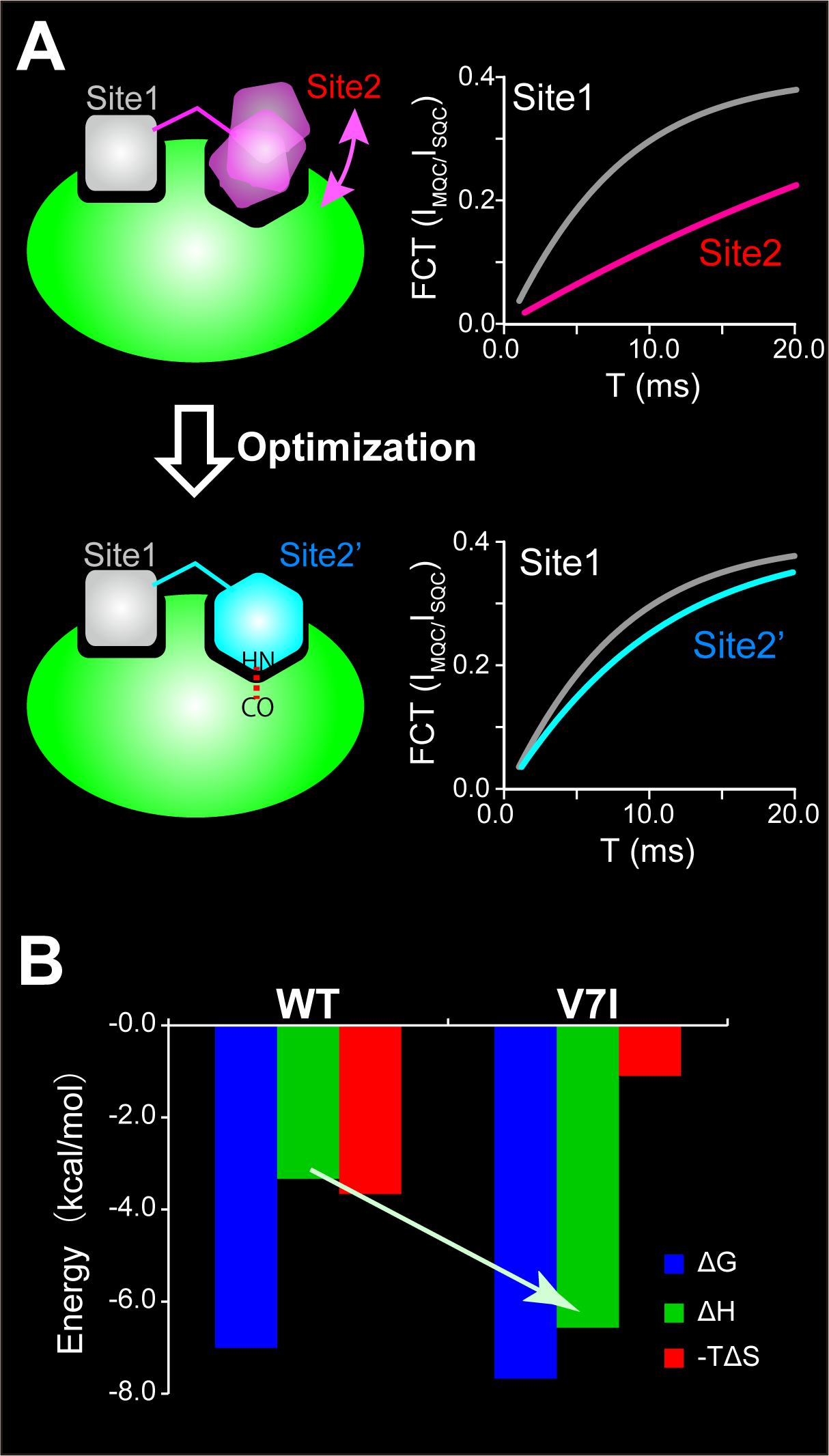
The thermodynamic properties of ligand-receptor interactions attract attention due to their relation to the binding specificity. However, designing ligands with preferable thermodynamic properties is challenging. We developed a strategy to experimentally evaluate the local dynamics and the surface complementarity of ligands in the receptor-bound state by forbidden-coherence-transfer (FCT) analysis and utilized the information to ligand optimization (Panel A). The FCT method was originally developed in L. Kay’s group, for protein dynamics analysis; however, we extended the method to dynamic ligand optimization (Angew. Chemie. Int. Ed., 2016; Molecules, 2017).
By applying the FCT method to the interaction between the MEF2A peptide and the p38α MAP kinase, the dynamics and surface complementarity of the MEF2A methyl groups in the p38α-bound state were quantitatively analyzed. Based on the information, we introduced a rational modification to the MEF2A peptide and obtained a ligand with higher affinity and better thermodynamic properties (higher enthalpic contribution for binding) than those of the original ligand (Panel B). The ligand is expected to show an improved specificity to its receptor, which is beneficial for the structure-guided lead-optimization.
Protein kinase regulation

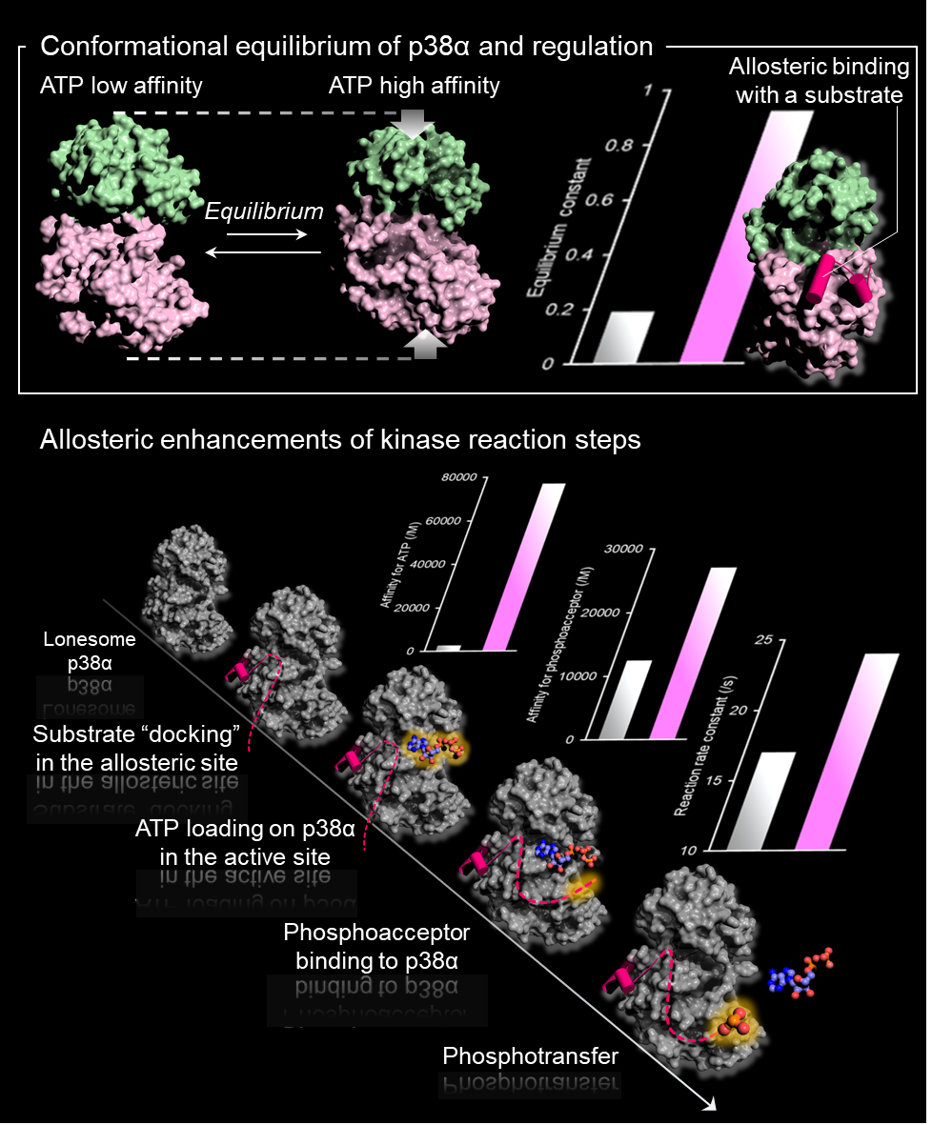
A protein kinase works in a signaling pathway or a molecular network via interactions with various proteins including substrates, upstream kinases, phosphatases, and scaffold proteins. The systems-biology approaches have been utilized for estimating and visualizing the behavior of signaling output. However, it is rarely considered that the molecular interactions and enzymatic activity could be modulated by the inherently dynamic nature of protein structures that reflect various physicochemical factors.
We think experimental elucidation of structure and dynamics of proteins are indispensable to analyze biological signaling. Toward this end, we revealed that a stress-responsive MAPK, p38α, fluctuates between conformational states with distinct affinities for ATP. Interactions with specific substrates at an allosteric site up-regulated the kinase activity of p38α by modulating the conformational equilibrium. This mechanism allows robust stress signaling, where p38α efficiently phosphorylates substrates even with a limited concentration of ATP under a stressed cellular environment (Nat. Struct. Mol. Biol., 2014). An integrative and comprehensive approach that takes the dynamic properties of protein structure into account, is one of our primary interests to capture the dynamic behavior of an entire signaling network.